
| Livestock Research for Rural Development 31 (8) 2019 | Guide for preparation of papers | LRRD Newsletter | Citation of this paper |
Cyprinus carpio is an important fish species in Albania, inhabiting natural lakes, pond, reservoirs and in hatcheries. Fingerlings produced in hatcheries are released for restocking of natural lakes and ponds. Four microsatellite markers were used to analyze genetic diversity of five carp populations from three natural lakes and two hatcheries in Albania. Genetic diversity between populations was moderate, with a mean value of 076, indicating that 7.6% of variation was between populations. A relatively high level of gene flow and a moderate level of genetic differentiation based FST values were found between all populations. Inbreeding value in whole population was high 45.8%. Structure analysis revealed three groups. These was supported by FCA and dendrogram based on Nei's genetic distance.
Keywords: genetic diversity, inbreeding, genetic structure, genetic distance, FCA
Albania is rich in water resources and common carp is a fish species that plays an important role for the local community regarding the socio economic aspect. Microsatellites as highly variable molecular markers are used for several purposes. (Tripathy et al 2018) report that microsatellites in fishery, are used for many purposes like, studying genetic variations of closely related species, pedigree analysis, identifying fish stock etc. for conservation strategies in fisheries and aquaculture management.
Microsatellites are used as molecular markers to genetically characterize Albanian carp population from natural lakes, Shkodra, Ohrid and Prespa (Biba et al 2014, Biba et al 2015, Biba et al 2015) as well as, from two hatcheries of Tapiza and Klosi (Bixheku et al 2019, Bixheku et al 2019)). The fingerlings produced in Tapiza and Klosi hatcheries are used for stocking ponds and lakes of Albania. Microsatellites are used for the estimation of genetic variability of natural and hatcheries carp populations from Croatia (Tomljanovic et al 2013), China (Zhou et al 2004), Caspian Sea (Ahmadi et al 2018), or even for three continents (Chen et al 2012). A previous study has analyzed genetic diversity of carp populations from natural lakes in Albania, based on microsatellite markers (Biba et al 2017). In the present study we extend the research carried out previously and intend to study the genetic variability within and among carp populations from natural lakes and hatcheries, by the use of four microsatellite markers. The main objective is to obtain information from natural and hatchery populations for fisheries management and conservation programs.
A total of 150 carp individuals from 5 populations were genotyped for 4 microsatellite loci, as described previously (Bixheku et al 2017). Individuals were sampled from three natural lakes: Ohrid, Shkodra, Prespa and two hatcheries: Tapiza and Klosi.
The observed number of alleles (NA), effective number of alleles (NE), observed (HO) and expected (HE) heterozygosity per locus and populations were determined using GenAlex 6.3 software (Peakall et al 2006). Allelic richness (AR), Mean Number of Alleles per locus (MNA), Wright F-statistics (FIS, F IT and FST) were calculated using FSTAT v 2.9.3 (Goudet 2001).
Exact tests for deviations from Hardy Weinberg equilibrium (HW) by breed were estimated with the software Genepop 4.2 (Raymond 1995), using Monte Carlo simulations via Markov chains.
Analysis of Molecular Variance was carried out by ARLEQUIN 3.1 (Excoffier et al 2005) in order to determine the partitioning of genetic variation between and within groups and populations. The significance levels were obtained by 1000 permutations. UPGMA dendrogram based on Nei’s genetic distance was constructed by POPULATIONS program 1.2.30 (Langella 2002). The bootstrap values were determined via 1000 replicate across loci. The dendrogram was visualized by TREEVIEW 1.6.6 software (Page 2001).
The patterns of the population structure were assessed using the Bayesian clustering approach in STRUCTURE 2.3 (Pritchard et al 2000) assuming admixture and independent allele frequencies, with k values from 2 to 5, where 5 runs were applied for each K.. A total of 20000 iterations were used as burn-in period for all runs. Data was collected during 10000 iterations. To determine the most likely hierarchical level of genetic structure, values of LnP(D) for each K and the estimated delta K (DK) were plotted (Evanno et al 2005). Graphic presentation of these statistics was obtained with the web-based Structure Harvester v0.6.8 (Earl et al 2012)
Factorial Correspondence Analysis (AFC) was performed to test the possible admixtures using GENETIX software (Belkhir 2004).
For the whole carp population a total of 287 alleles were found for 150 genotyped individuals for 4 microsatellite markers. Gene diversity indices for five populations are shown in Table 1 and Table 2.
|
Table 1. Genetic variability of 5 populations at 4 microsatellite loci |
|||||
|
Pop |
MFW1 |
MFW6 |
MFW7 |
MFW18 |
|
|
Ohrid |
Na |
23 |
19 |
22 |
20 |
|
Ne |
11.842 |
12.519 |
12.376 |
9.921 |
|
|
I |
2.758 |
2.712 |
2.792 |
2.637 |
|
|
Ho |
0.633 |
0.731 |
0.600 |
0.320 |
|
|
He |
0.916 |
0.920 |
0.919 |
0.899 |
|
|
uHe |
0.931 |
0.938 |
0.938 |
0.918 |
|
|
Shkodra |
Na |
26 |
33 |
25 |
19 |
|
Ne |
17.357 |
23.041 |
17.566 |
8.191 |
|
|
I |
3.041 |
3.320 |
3.028 |
2.558 |
|
|
Ho |
0.407 |
0.793 |
0.296 |
0.481 |
|
|
He |
0.942 |
0.957 |
0.943 |
0.878 |
|
|
uHe |
0.960 |
0.973 |
0.961 |
0.894 |
|
|
Prespa |
Na |
16 |
15 |
19 |
11 |
|
Ne |
9.058 |
8.503 |
11.879 |
3.122 |
|
|
I |
2.448 |
2.421 |
2.697 |
1.653 |
|
|
Ho |
0.160 |
0.462 |
0.286 |
0.333 |
|
|
He |
0.890 |
0.882 |
0.916 |
0.680 |
|
|
uHe |
0.908 |
0.900 |
0.932 |
0.693 |
|
|
Klosi |
Na |
14 |
30 |
22 |
15 |
|
Ne |
5.568 |
18.233 |
9.941 |
8.494 |
|
|
I |
2.088 |
3.170 |
2.709 |
2.405 |
|
|
Ho |
0.522 |
0.821 |
0.462 |
0.632 |
|
|
He |
0.820 |
0.945 |
0.899 |
0.882 |
|
|
uHe |
0.839 |
0.962 |
0.917 |
0.906 |
|
|
Tapiza |
Ne |
3.982 |
19.882 |
4.017 |
9.116 |
|
I |
1.632 |
3.158 |
1.738 |
2.618 |
|
|
Ho |
0.333 |
0.808 |
0.364 |
0.429 |
|
|
He |
0.749 |
0.950 |
0.751 |
0.890 |
|
|
uHe |
0.775 |
0.968 |
0.768 |
0.906 |
|
|
Table 2. Mean allelic pattern across populations |
|||||
|
Pop |
He |
Ho |
MNA |
Ar |
FIS |
|
Ohrid |
0.913 |
0.571 |
21.00 |
15.550 |
0.376 |
|
Shkodra |
0.930 |
0.495 |
25.75 |
18.312 |
0.469 |
|
Prespa |
0.842 |
0.310 |
15.25 |
12.293 |
0.624 |
|
Mean (Natural) |
0.895 |
0.459 |
20.667 |
15.385 |
0.490 |
|
Klosi |
0.887 |
0.609 |
20.25 |
15.134 |
0.316 |
|
Tapiza |
0.835 |
0.483 |
16.50 |
12.896 |
0.435 |
|
Mean (Hatchery) |
0.861 |
0.546 |
18.375 |
14.015 |
|
|
Mean (All Populations) |
0.881 |
0.494 |
19.75 |
14.837 |
0.444 |
|
He: expected heterozygosity; Ho: Observed
heterozygosity; MNA: Mean number of |
|||||
The average value of expected heterozygosity for the whole population was 0.881, indicating that the whole carp population has a high genetic diversity. The average observed heterozygosity value was 0.494 for the whole population. In all cases the HE values were higher than H O.
Wright's fixation index (FIS) was 0.458, indicating a high level of inbreeding. Also all markers showed high positive values, indicating an excess of heterozygotes deficit. The FIT index, which show the heterozygosity loss of an individual with respect to the whole population showed a lack of heterozygotes of 49.9%. The FST index that measures the degree of genetic diversity showed that the differences between populations were 7.6% and differences between individuals were 92.4% (Table 3). The results indicate that the genetic differentiation between populations was low.
|
Table 3. F statistics estimates over all populations for each loci |
|||
|
Locus |
FIS |
FIT |
FST |
|
MFW1 |
0.528 |
0.567 |
0.082 |
|
MFW6 |
0.237 |
0.263 |
0.033 |
|
MFW7 |
0.568 |
0.599 |
0.071 |
|
MFW18 |
0.509 |
0.562 |
0.110 |
|
Total |
0.458 |
0.499 |
0.076 |
Hardy Weinberg equilibrium testes per population and locus showed significant deviation (p< 0.05) in all 20 tests performed. This is due to heterozygote deficit in each population.
Estimated FIS values per each population were high indicating deficit of heterozygotes per each population. This can be explained by inbreeding, selection, the presence of population substructure and presence of null alleles.
UPGMA dendrogram based on NEi's standard genetic distance showed three different clusters (Figure 1). Prespa is separated from the others. In the second cluster were Ohrid and Shkodra and the third cluster were hatchery populations. Bootstrap values at the nodes of the tree were higher than 50, showing robustness of the tree.
 |
| Figure 1. UPGMA dendogram based on DA distance for 5 populations of common carp |
The population structure was analyzed by the use of STRUCTURE program. According to Evanno method (Evanno et al 2005), implemented in the Structure Harvester software (Earl et al 2012), it was assumed that k = 3 is the most likely number of ancestral populations that contribute to the genetic diversity of Albanian carp (Figure 2). Results show a clear differentiation of populations from natural lakes and hatchery. The proportion of membership is shown in table 4. The structure analysis supported FCA, with three genetically distinct population, where Ohrid and Shkodra were grouped together, Klosi and Tapiza hatchery grouped together and Prespa. The FCA as shown in figure 3, reveals three group of populations.
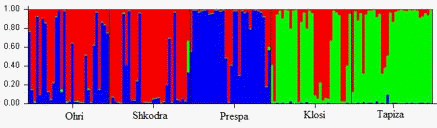 |
| Figure 2. Population structure estimated with STRUCTURE in Albanian carp populations for K =3 |
|
Table 4. Proportion of membership for each of the five |
|||
|
Groups |
Cluster |
||
|
1 |
2 |
3 |
|
|
Ohri |
0.570 |
0.008 |
0.422 |
|
Shkodra |
0.774 |
0.017 |
0.209 |
|
Prespa |
0.168 |
0.007 |
0.826 |
|
Klosi |
0.396 |
0.597 |
0.007 |
|
Tapiza |
0.142 |
0.847 |
0.011 |
 |
| Figure 3. The FCA for 5 carp populations |
To explain the partition of genetic variation within and between population AMOVA analysis (Excoffier et al 2005) was performed. We grouped the populations according to the branches observed in the phylogenetic trees. Results of AMOVA analysis are shown in table 5. The results shows that 49.832% of the total variation came from within individuals. Variation among populations within groups was 5.385%
The gene flow between hatchery populations was very limited (Table 6). Also the genetic differentiation between populations was moderate between all pairs of populations, except of Ohrid and Shkodra populations
|
Table 5. AMOVA analysis results for all populations of common carp |
|||
|
Source of variation |
Sum of |
Variance |
Percentage |
|
Among groups |
24.98 |
0.054 |
2.723 |
|
Among populations within groups |
15.75 |
0.107 |
5.385 |
|
Among individuals within populations |
324.234 |
0.834 |
42.057 |
|
within individuals |
127.000 |
0.989 |
49.833 |
|
Total |
491.963 |
1.985 |
|
|
Table 6. FST values (upper diagonal) and gene flow (below diagonal) among all carp populations |
|||||
|
Ohri |
Shkodra |
Prespa |
Klosi |
Tapiza |
|
|
Ohri |
0.022 |
0.054 |
0.072 |
0.095 |
|
|
Shkodra |
11.07 |
0.061 |
0.064 |
0.087 |
|
|
Prespa |
4.42 |
3.82 |
0.109 |
0.131 |
|
|
Klosi |
3.20 |
3.68 |
2.04 |
0.099 |
|
|
Tapiza |
2.37 |
2.61 |
1.67 |
2.28 |
|
Genetic variability and structure, population differentiation and effective population size of the different carp populations from natural lakes and hatchery are analyzed. These are important indicators for sustainable utilization of genetic resources common carp. Anyway, we recommend the use of a larger number of markers for fine scale genetic characterization. Population genetic structure analysis of important fish species is essential for optimizing management strategy or a stock improvement program (Islam et al 2007).
The results presented here show a decline of the observed heterozygosity relative to expected heterozygosity in all populations. Observed heterozgosity values for natural and hatchery carp populations were lower compared with Greek populations (Karaiskou et al 2011), but similar with values found in Croatia (Tomljanovic 2013), or from France region and Czech Republic (Desvignes et al 2001). On the other hand we found increased inbreeding in all populations (FIS -0.333-0.643), with a mean value of 0.458.
STRUCTURE analysis, AMOVA and UPGMA dendrogramm indicated differentiation between natural population and hatchery. Quite low was the gene flow also between Prespa and hatcheries populations, explaining also the separate group and differentiation of carp population from Prespa natural lake. The highest pairwise FST values were between Prespa and hatcheries population, followed by values between hatcheries populations, indicating differences between these populations (Table 6). The overall genetic differentiation is moderate, where the variation between population was 7.6%.
According to Singh et al, (Singh et al 2011) the microsatellite technique opens new perspective for studying the structure of closely related populations, population samples over a reduced geographic scale and less isolated populations, therefore being suitable for these populations under study. Expected heterozgosity and the number of alleles are used as indicators of genetic diversity (Desvignes et al 2001). MNA and expected heterozygosity values were higher in natural populations than in hatcheries. (Tomljanovic 2013) found also a higher genetic variability of the feral carp populations in comparison to the hatchery stocks in Croatia. (AHMADI et al 2018) reported that allelic and gene diversity of carp hatchery populations tended to be lower compared to the wild populations. Genetic variability in hatchery stocks and natural populations has been reported for many species. (Li et al 2017) found that in Hemibarbus maculates in aquacultured populations exhibited high genetic distances and significant genetic differentiation compared to the wild population. (Kashiri et al 2018) found in Rutilus kutum a little lower level of allelic diversity in the hatchery population compared to the wild populations. (Tomljanovic 2013) suggest that the reduction of allelic diversity in hatchery stocks might be the result of founder events or occasional bottleneck effects during the breeding process.
The results obtained here regarding genetic variability and relationship within and among natural and hatcheries carp populations will help in designing conservation and breeding programs.
Ahmadi M, Kashiri H, Shabani A And Moghadam A A 2018 Genetic variability in wild and hatchery populations of commercially important fish: The common carp (Cyprinus carpio). Biodiversitas Journal of Biological Diversity. 19, (4): 1468-1474.
Belkhir K 2004 1996-2004 GENETIX 4.05, logiciel sous Windows TM pour la genetique des populations. genetix/genetix/genetix. htm.
Biba A, Hoda A and Vardhami E 2014 Genetic diversity of Cyprinus carpio from Ohrid Lake, estimated by four microsatellite loci. Albanian Journal of Agricultural Sciences. 253.
Biba A, Hoda A and Vardhami E 2015 Genetic diversity of Cyprinus carpio of Prespa Lake based on microsatellite loci. Bul Shkenc Nat. 19, 12-19.
Biba A, Hoda A, Bozgo V and Mali S 2017 Genetic diversity of Cyprinus carpio of natural lakes in Albania estimated by microsatellite loci. Endocytobiosis & Cell Research. 28 , (1):.
Biba A, Hoda A, Vardhami E and Bozgo V 2015 Genetic diversity of common carp from Shkodra Lake based on microsatellite markers. Livestock Research for Rural Development. 27 (4): 3-5.
Bixheku X, Hoda A and Bozo D 2017 Allelic frequencies of MFW7 microsatellite loccus in carp (Cyprinus carpio) of two fish farming centers. Albanian Journal of Agricultural Sciences. 19-21.
Bixheku X, Hoda A and Bozo D 2019 Allele Frequency Data of 4 Microsatellite Markers of Cyprinus carpio from Tapiza Hatchery in Albania. International Journal of Agriculture Innovations and Research. 7, (4): 455-457.
Bixheku X, Hoda A and Bozo D 2019 Genetic diversity of Cyrpinus carpio from an Albanian fish hatchery based on microsatellite markers. Acta Biologica Turcica. 32, (3): 112-116.
Chen Q, Wang C, Lu G, Zhao J, Chapman D C, Zsigmond J and Li S 2012 Microsatellite genetic diversity and differentiation of native and introduced grass carp populations in three continents. Genetica. 140, (4-6): 115-123.
Desvignes J F, Laroche J, Durand J D and Bouvet Y 2001 Genetic variability in reared stocks of common carp (Cyprinus carpio L.) based on allozymes and microsatellites. Aquaculture. 194, (3-4): 291-301.
Earl D A and VonHoldt B M 2012 STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation genetics resources. 4, (2): 359-361.
Evanno G, Regnaut S and Goudet J 2005 Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular ecology. 14, (8): 2611-2620.
Excoffier L, Laval G and Schneider S 2005 Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary bioinformatics. 1, 117693430500100003.
Goudet J 2001 FSTAT, a program to estimate and test gene diversity and fixation indices (version 2.9. 3). http://www2. unil. ch/popgen/softwares/fstat. htm.
Islam M N, Islam M S and Alam M S 2007 Genetic structure of different populations of walking catfish (Clarias batrachus L.) in Bangladesh. Biochemical genetics. 45, (9-10): 647-662.
Karaiskou N, Lappa M, Kalomoiris S, Oikonomidis G, Psaltopoulou C, Abatzopoulos T J, Triantaphyllidis C and Triantafyllidis A 2011 Genetic monitoring and effects of stocking practices on small Cyprinus carpio populations. Conservation genetics. 12, (5): 1299-1311.
Kashiri H, Shabani A, Gorgin S, Rezaii M and Jabale A R 2018 Microsatellite DNA Marker Analysis of Genetic Variation in Wild and Hatchery Populations of Caspian Kutum (Rutilus kutum). Turkish Journal of Fisheries and Aquatic Sciences. 18, (9): 1101-+.
Langella O 2002 Populations, a free population genetic software. URL http://www. legs. cnrs-gif. fr.
Li L, Lin H, Tang W, Liu D, Bao B and Yang J 2017 Population genetic structure in wild and aquaculture populations of Hemibarbus maculates inferred from microsatellites markers. Aquaculture and Fisheries. 2, (2): 78-83.
Page R D 2001 TreeView. Glasgow University, Glasgow, UK.
Peakall R and Smouse P E 2006 GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular ecology notes. 6, (1): 288-295.
Pritchard J K, Stephens M and Donnelly P 2000 Inference of population structure using multilocus genotype data. Genetics. 155, (2): 945-959.
Raymond M 1995 GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248-249.
Singh A, Katiyar P, Yadav M and Tiwari A 2011 Microsatellites: simple sequences with dynamic role in fish genetics. Intern. J. Biomed. Res. 2, 123-137.
Tomljanovic T, Treer T, Cubric V, Safner T, Sprem N, Piria M, Matulic D, Safner R and Anicic I 2013 Microsatellite-based genetic variability and differentiation of hatchery and feral common carp Cyprinus carpio L.(Cyprinidae, Cypriniformes) populations in Croatia. Archives of Biological Sciences. 65, (2): 577-584.
Tripathy S K and others 2018 Broad Spectrum Utilities of Microsatellite in Fish and Fisheries. Journal of Biotechnology Research. 4, (6): 29-45.
Zhou J, Wu Q, Wang Z and Ye Y 2004 Genetic variation analysis within and among six varieties of common carp (Cyprinus carpio L.) in China using microsatellite markers. Russian Journal of Genetics. 40, (10): 1144-1148.
Received 2 July 2019; Accepted 8 July 2019; Published 1 August 2019